Genomics of Adaptation
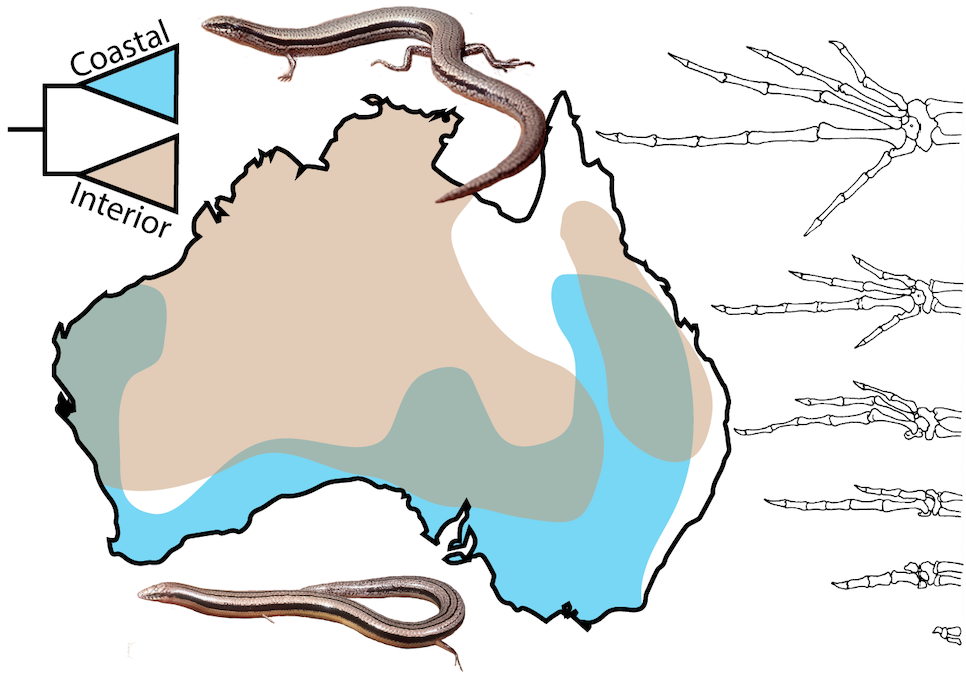
Since beginning as a researcher, I have been interested in understanding the process of adaptation, particularly at the level of genetics. During my Ph.D., I used combinations of genomic data to examine rapid adaptation to novel ecological conditions in invasive Florida Burmese pythons (Card et al., 2018, Molecular Ecology) and the repeated evolution of island dwarf phenotypes in Central American boa constrictors (Card et al., 2019, Genome Biology & Evolution). My postdoctoral research is focused on understanding interspecies differences in limb morphology in the Australian skink genus Lerista, where species vary from full pentadactyl limbs to total limblessness with several intermediate forms. As part of this project, I am combining morphological and whole-genome data for dozens of species with functional genomic data collected from embryos to provide an integrated look into the genetic and molecular underpinnings of limb morphology.
Convergent Evolution
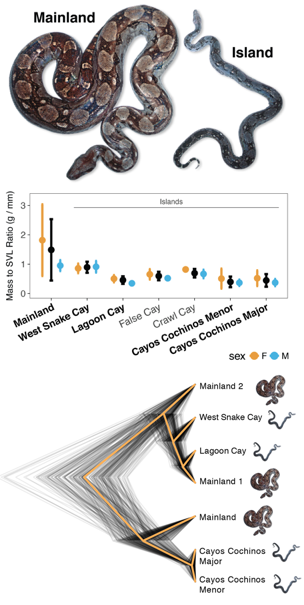
Convergent evolution provides some of the strongest evidence for the process of evolution by natural selection. My research has focused on adding to the small, but growing, body of literature describing systems where phenotypic convergence is mediated by molecular convergence. My doctoral research examined repeated evolution of island dwarf phenotypes in Central American boa constrictors (Card et al., 2016. MPE; Card et al., 2019, Genome Biology & Evolution). My current research focusing on the evolution of reduced limb morphologies in Australian Lerista also provides the opportunity to examine shallow-scale convergence, as limb morphotypes have evolved independently in two Lerista clades.
Genome Structure, Function, & Evolution
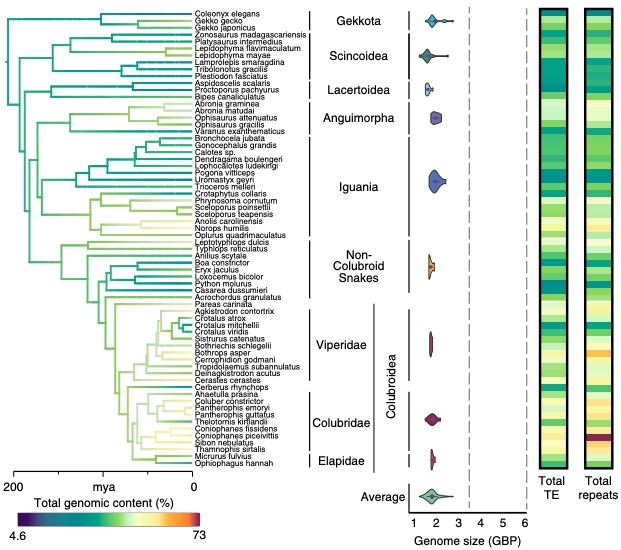
The structure of genomes, including overall size, karyotype, repetitive content, and more, is an important driver of organism phenotype and evolves in interesting ways. As a researcher, I have contributed to producing several genomic resources for squamate reptiles, including the Burmese python - the first snake genome (Castoe et al., 2013, PNAS), a genome for the garter snake - an important model organism in ecology (Perry/Card et al., 2018, Genome Biology & Evolution), and the prairie rattlesnake - the first chromosome-scale assembly for any squamate (Schield et al., 2019, Genome Research). I have also contributed to work seeking to understand lineage-specific patterns of repetitive content, including microsatellite content across vertebrates (Adams et al., 2016, Genome) and repetitive landscapes across squamate reptiles (Pasquesi et al., 2018, Nature Communication).